Complete bacterial diversity profiling (bactDNA) is part of our suite of NGS analyses and is used to provide clarity on the diversity and composition of the bacterial community present in rivers, lakes, bores, reservoirs, sewage, treated water, in short - any water matrix. The test provides a ranking of the presence of bacterial DNA from highest to lowest prevalence (as a percentage of the total DNA extracted from the sample).
Conventional methods for detecting bacterial contamination are based on traditional culture techniques which only target specific bacteria, some of which require days of incubation. This means scientists only identify those bacteria they are searching for – while others are left out.
The benefits of Next Generation Sequencing (NGS) of bacteria can be realised without the need for any of the traditional culture techniques. All that is required is a 1L water sample (or 1g of soil) from which we screen the microbial community genomes present.
Our technique enables us to identify strains of bacteria that usually cannot be successfully cultured, take long periods of time to grow and/or require complicated growth techniques to get a result.
A quantum leap in testing for bacterial contamination
Bacterial Diversity Profiling enables us to identify the relative proportion of micro-organisms present in a mixed microbial community. This detailed snapshot of the microbial community can have many applications, for example to examine taste and odour issues in water due to biofilms, to detect the presence of problematic organisms in surface water, in groundwater bores or to assess and track the health of microbial populations in wastewater treatment plants for increasing the efficiency of operational processes.
Furthermore, the bacterial DNA data can be used to make assessments over time, based on sample baseline data. The bacterial DNA detected in a sample can be monitored to determine if the community structure is shifting to favour different bacterial groups and becoming more problematic. The bacterial DNA diversity profile represents the current status of the sample and should remain relatively static, depending on the source, volume and (if applicable) disinfection protocols.
Results
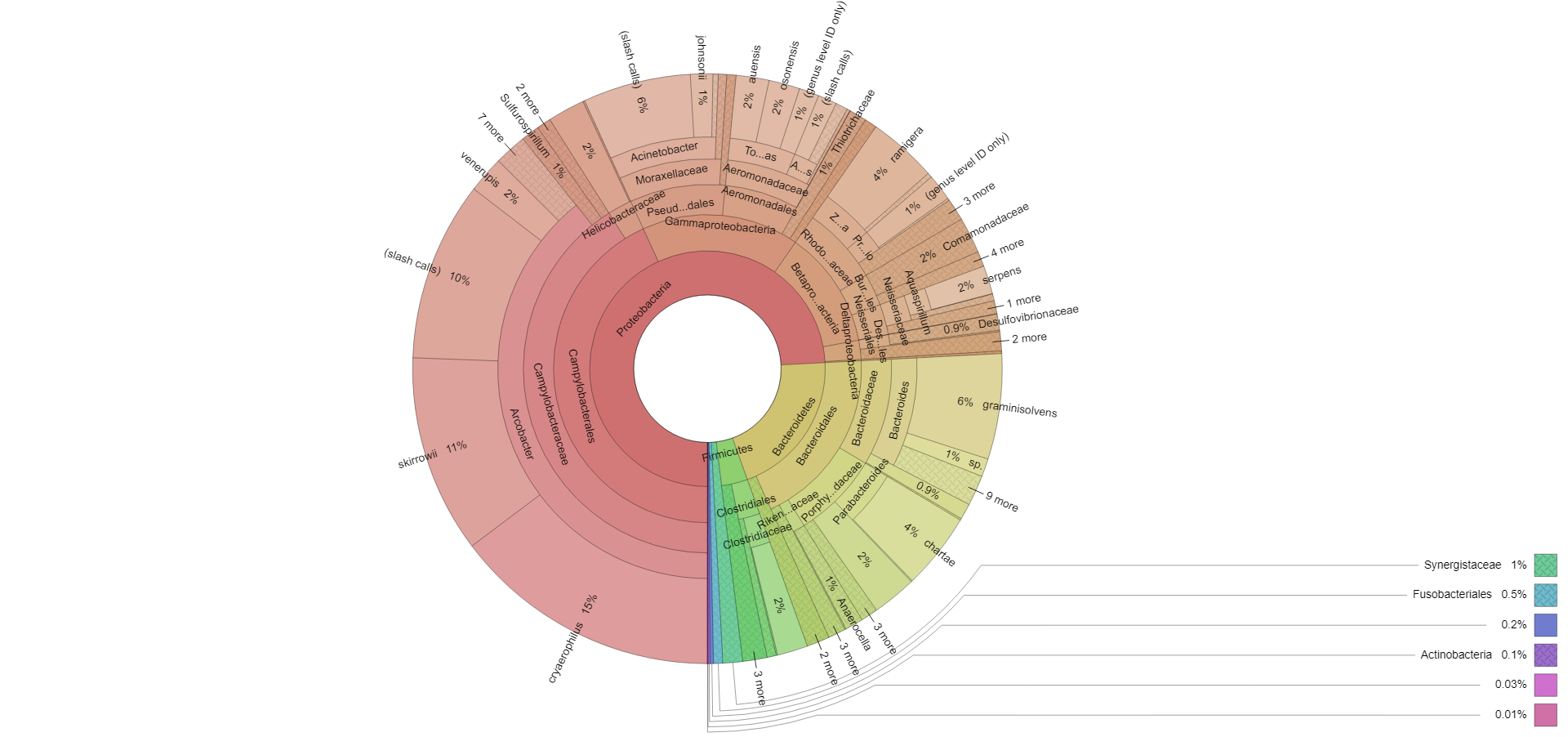
Results are presented in a NATA accredited, user-friendly report (PDF format) which includes a ranking of bacteria in order of relative abundance (% of mapped reads). The report also includes dot-point comments against orders detected.
Interpretational reporting in pdf/graphic formats allowing characterisation of the bacterial population, taxonomical analysis and species identification in the supplied sample. This is further enhanced through visualisation in the report. Interactive Krona plots are also provided to enable ‘zoomable’ visualisation of your results. Operational Taxonomic Unit (OTU) tables and raw sequencing files are available on request.
Our application provides the most extensive 16S rRNA coverage currently available in the Australian water industry. Our method examines seven of the nine hypervariable regions in the bacterial 16s rRNA gene - currently the most of any system and all in one efficient process. Our process compares the sequences against our defined and curated database, thus delivering the most discriminatory and sensitive bacterial NGS system available. If required, any unknown isolates can be further identified, thus creating a unique database.
Samples are PCR amplified, with universal primer sets covering seven of the nine hypervariable regions of bacterial 16S rRNA. This enables the identification of a broad range of bacteria down to the genus/species level. The amplicons are barcoded, pooled and then sequenced. Proprietary bioinformatics software and curated reference databases are used to identify the bacteria present in samples.
Using this technology, we are able to deliver the most discriminatory and sensitive bacterial NGS system available. The AWQC method employs two pieces of robotic equipment - an ION Chef(TM) and an ION S5(TM) - to create DNA chips and unique barcodes for the bacteria found in a water sample, providing detailed and reliable information.
With three independent bacterial bioinformatics pipeline methods on offer (custom BLAST, QIIME2 and Kraken2), we can customise our analysis for any bacterial issue encountered, including complex biofilms.
This analysis provides semi-quantitative abundance estimates of the organisms prevalent in a sample by percentage and as such provides a means to compare microbial populations between samples.
Note: The size of the water body, habitat of the defined species, time of year and period of collection, water flow and other hydrological factors need to be taken into consideration for optimal sampling design and subsequent bacterial diversity profiling to be effective.
Sample container
Water matrix: two sterile, DNA free, sodium thiosulphate dosed 1L PET bottles
Sludge / soil / sewage matrix: minimum of 1g in sterile sludge pot/container
Label
1.25L - Bacto – ptdna – none - sterile – air gap
Analytes & holding times
All water types
Sludge / soil / sewage matrix
Process within a maximum of 24hrs from time of collection.
Notes
Sampler must follow DNA sampling procedure WI-375
Turnaround Time (TAT)
* The DNA from all samples is extracted on receipt and stored until analysis. AWQC schedules one bacterial diversity profiling run in the third week of each month.
* By special request, samples can be run as soon as possible on receipt.
